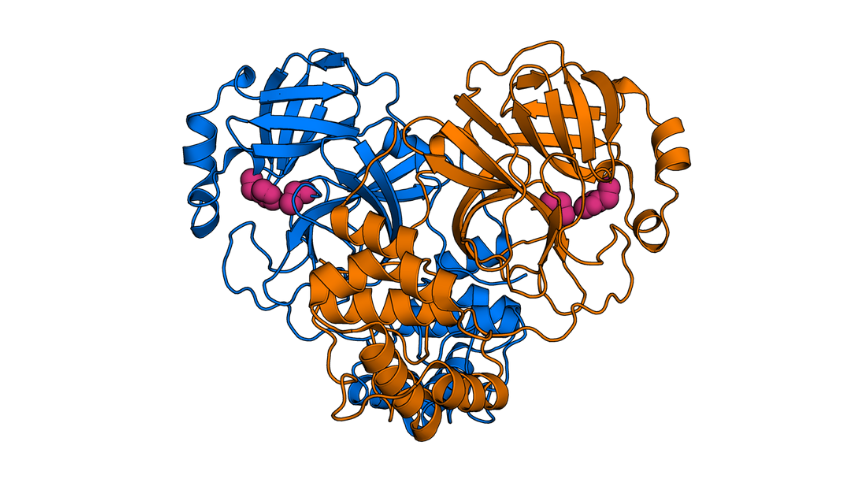
Image: The protease is the target of the COVID-Moonshot project, a collaboration F@h is participating in with chemists and biologists around the world. Read more about the project, and see it featured on F@h’s “Meet the Proteins” series.
As the COVID-19 pandemic makes waves around the world, scientists and researchers are searching for a remedy. One project, Folding@home (F@h), has emerged as a leader in this research. Launched in 2000, F@h is a distributed computing project through which citizen, or volunteer, scientists study and simulate proteins. Since its launch, the project has played a role in research on Alzheimer’s disease and Ebola. Now, participating scientists are tackling COVID-19.
We connected with Sukrit Singh (Ph.D. candidate, Washington University in St. Louis; and, F@h team member) to learn more about the project’s background, its impact to date on COVID-19 research, and the importance of the citizen-scientist community. Also, John Fenno (global marketing director of GeForce, NVIDIA) shares why the team at NVIDIA has long been a collaborator on the project.
“Folding@home has been doing great work for many years, rallying gamers to donate extra computing capacity for research. When COVID-19 hit California, we were scared and nervous like everyone else, and our social media team asked what we could do to help,” Fenno says. “At the same time, Pedro at PCMR alerted us to renewed F@h efforts, supporting research. F@h needed some help driving awareness — it was a no-brainer for us to promote the effort globally through our GeForce social channels.”
Read on for a deep dive into Folding@home from Singh.
SIGGRAPH: Share some background on Folding@home. How did it get its start? How has it supported past disease research before taking on coronavirus?
Sukrit Singh (SS): Folding@home (F@h) launched in 2000 as a distributed computing project to study “protein folding,” the phenomena by which the proteins in our body (linear chains of amino acids) assemble into complex shapes that allow us to see, move, think, and more. Imagine if you were able to take the individual parts of a car and lay them out in your driveway, and once you did, the components spontaneously assembled themselves into a fully functioning car!
Computers can simulate the movement of atoms over time with an approach known as molecular dynamics (MD). MD acts as a “computational microscope” that lets us study atomic motions and how they relate to protein behavior and biological function. However, studying protein folding is a computationally expensive task, and generating sufficient data to study protein motions can take hundreds of years on a single desktop computer.
Inspired by SETI@home, F@h founder Vijay Pande and his team at Stanford built F@h as a way to run MD simulations of proteins. To achieve this, the team worked out a way to break up MD simulations into smaller chunks and distribute them across the internet. This way, the protein motions that might take hundreds of years to observe on a single desktop can be modeled in much less time by distributing the calculation across hundreds of desktops. In 2019, Prof. Greg Bowman (Washington University in St. Louis) became director of the F@h project.
Many early breakthroughs were made over the next decade with F@h, including the first observation of the complete folding pathway of a protein in 2010. Over 200 papers using F@h have been published on a variety of topics. Notably, F@h simulations have allowed scientists to find novel, small-molecule inhibitors to β-Lactamase and model never-before-seen processes in our cells that relate to cancer.
Most recently, the Bowman lab released a preprint that computationally identified a cryptic pocket in an ebolavirus protein that is critical for function. In the publication, this prediction is followed up with experiments that support the existence of this pocket.
Now, we are bringing all the power behind F@h to fight the coronavirus.
SIGGRAPH: Let’s get technical. How do citizen scientists use F@h?
SS: More information about the background of F@h and how the gaming community can help now can be found in here.
In terms of a description of what happens when you participate, here’s a brief overview: When someone downloads the F@h app and runs our software (the “client”), they are connected to our servers and receive a “work unit”, which is one part of a simulation. Each simulation is a collection of atoms moving over time. The movement of these atoms relative to one another can be described using Newton’s laws of motion. Over a long enough simulation, the proteins will “shapeshift” and change their structure. These new structures can have useful information, explaining biological functions that the protein performs or providing insight into why certain mutations cause disease. Some new structures may even present new druggable opportunities.
Each simulation, which starts from a single configuration of atoms with positions and velocities, is broken up into work units with each one representing one temporal component of the whole simulation. This means that a completed work unit has “time-series” data of atoms and their positions over time. Each work unit is completed sequentially — that is, one work unit must be completed, and the end of that work unit is the start of the subsequent work unit for the next client.
SIGGRAPH: How do you fold proteins?
SS: When we analyze work units that our community returns, we do so by stitching the work units together into long time-series trajectories. For any one project, we end up with multiple “movies” of the atoms moving over time and the protein shape-shifting.
We then utilize multiple statistical methods to parse and analyze these “movies”. In particular, we often construct Markov state models (MSMs), which are “map” representations of a protein’s structural diversity. In these MSMs, each “state” is a unique structure of the protein where the atoms are in a specific, distinct configuration. We then use F@h data to estimate the probability of moving from any one state into another. This way, we have a view of the full “landscape” of structures a protein can adopt.
One particularly useful feature of these MSMs is our ability to identify new areas on proteins that can serve as targets for drug design. As the protein moves over time (adopting different states in our MSM), the atoms shift around relative to one another which exposes new surfaces, grooves, or pockets of atoms to the environment. These previously unobserved “cryptic pockets” often are appropriately shaped to bind a drug-like molecule and present promising targets for future drug design methods.
These cryptic pockets can be hard to identify with experimental data alone, but they can be observed using the computing power of F@h. In fact, as I mentioned above, we recently published a paper where we utilized F@h and our computational toolset to identify a cryptic pocket in an ebolavirus protein and were able to experimentally support its existence. Importantly, jamming this pocket open with a small-molecule label inhibits protein function, making this pocket a promising target for drug design methods! This paper is available currently as a preprint on bioRxiv.
SIGGRAPH: How has Folding@home impacted COVID-19 so far?
SS: It is important to note that while scientific research takes time, we have already made progress and new insights into COVID-19 as we have scaled up the research infrastructure of F@h!
One particularly exciting finding we recently released was our observation of the motions of the SARS-CoV-2 Spike protein. The Spike protein exists on the surface of the virus (visible in most renderings of the viral particle) and undergoes an opening process that allows it to bind human host cells to infect them. Using adaptive-learning algorithms and F@h, we have observed the process of opening. Knowing the states adopted in this opening process could be useful for antibody and therapeutic design.
We also are simulating proteins of SARS-CoV-2 to hunt for new cryptic pockets. Empowered by our community’s size and enthusiasm, we have launched almost every known protein structure from SARS-CoV-2 onto F@h, and, with our citizen scientists, we are generating data at a breakneck pace (greater than 100 times what we were generating prior to the pandemic). In the last few weeks, we have launched 126 projects focused on SARS-CoV-2 and almost 12 are already being run through our analysis pipelines to hunt for therapeutic opportunities.
Thanks to our network of citizen scientists, the combined compute power of F@h recently crossed into the exascale, climbing as high as 2.4 x 86 exaFLOPs. This makes us 15 times faster than IBM Summit and faster than the top 500 supercomputers in the world combined! This affords us the opportunity to try multiple strategies against this virus.
One such strategy F@h is participating in is the “COVID-moonshot,” a crowdsourced chemistry initiative to identify chemical inhibitors against one target of SARS-CoV-2, the main protease. Here, F@h simulations are simulating potential small-molecule inhibitors against the protease target, and this information is helping chemists around the world prioritize and optimize their synthetic strategies to design a viable therapeutic. These suggestions are crowdsourced, and anybody is welcome to submit ideas for new molecules to try out.
One awesome part of F@h has been watching communities, both academic and corporate, come together against this virus. We have been lucky to partner with academic groups around the world, such as the Joint European Disruptive Initiative (JEDI) and the COVID Moonshot project, as well as companies like Microsoft, Avast, NVIDIA, and Oracle. We also are thankful for the long-time support of many communities on the internet, such as Linus Tech Tips and the PCMR subreddit community.

Image credit: Folding@home team members during a Consortium Meetup in October 2019
SIGGRAPH: With over 1 million devices now part of F@h (even bitcoin miners have joined the charge), how do you continue to build the community? Why is the community aspect so important?
SS: The power of F@h is in its distributed nature and derives directly from our community. This means that every citizen scientist has been equally important in pushing F@h into exascale computing power and allowing us to tackle science at an unprecedented scale.
We would not be where we are now without the members of our community who have stepped up and helped out. We have grown almost 60 times in power and demand for work units, and with that we have gained many new users who may need support with the client or want to learn more about the science. With time and resources being limited, the long-time members of our community on the forums and our Discord channel have stepped up and started answering the hundreds of questions that have poured in. Our community members should feel proud for everything they’ve done to help us.
Further, this surge in demand and resources has given the F@h team more opportunities to interact with our community. Along with our forums and social media channels, we now have a verified Discord channel. This Discord channel is particularly exciting because community members can interact with one another, get support, compare Folding PC specs, or just generally hang out with their fellow citizen-scientists as we tackle COVID-19 together.
SIGGRAPH: Where do you envision F@h will be in the future, post-COVID-19?
SS: COVID-19 is just one of the many biomedical problems we are tackling, and we are excited to tackle more diseases with our newfound power. Prior to COVID-19, F@h simulations studied multiple biomedical problems from multiple types of cancer to antibiotic resistant infections. We want to bring the same power we have now against all of these maladies, working to identify new therapeutic strategies and making insight into fundamental biological questions.
We hope to retain many of our community members moving forward. This surge has been incredible, and we are developing ways to further engage with our citizen scientists to share progress and results as we generate data. Good science takes time, but we are excited to share all we have! We plan to release our data, methods, and code as open-source, and to publish on open-access websites like bioRxiv.
Further commenting on NVIDIA’s support of F@h, Fenno noted, “Our first tweet reached millions of gamers, and we’re happy to see that this and other ecosystem efforts helped mobilize gamers to renew their support for a great cause. Today, over 500K GeForce gamers are contributing their hardware, and F@h has achieved 2.5 exaFLOPs of power, making it the largest distributed supercomputer solution ever created. We’re proud of the whole gaming community for stepping up to support Folding@home. We can only hope the research efforts will help the medical community discover treatments as quickly as possible.”
 Sukrit Singh is a Ph.D. candidate in molecular biophysics at Washington University in St. Louis and a member of Prof. Greg Bowman’s lab and the Folding@home team. He started his thesis work in 2015, joining Prof. Bowman’s lab right after it had first opened. Since then, Sukrit has been involved in Folding@home primarily as a scientist and researcher but also has been managing social media, communications, and outreach since 2019.
Sukrit Singh is a Ph.D. candidate in molecular biophysics at Washington University in St. Louis and a member of Prof. Greg Bowman’s lab and the Folding@home team. He started his thesis work in 2015, joining Prof. Bowman’s lab right after it had first opened. Since then, Sukrit has been involved in Folding@home primarily as a scientist and researcher but also has been managing social media, communications, and outreach since 2019.
Prior to Folding@home’s shift to COVID-19 studies, Sukrit’s thesis work focused on studying information transfer in proteins, a phenomena known as “allostery.” Sukrit’s focus on allostery and protein dynamics centers around cell signaling, specifically on understanding molecular switches implicated in uveal melanoma and other cancers.
Sukrit originally hails from India, but also spent 10 years of his life in Singapore before obtaining his Bachelor’s degree in chemistry and biology (double major) at Washington University in St. Louis. During his undergraduate years, he worked in the lab of Dr. Garland Marshall, developing new peptidomimetics against HIV and bacterial infections.
 John Fenno is the global marketing director of GeForce at NVIDIA. He manages a shared resource team covering social media, influencer marketing, and original content videos.
John Fenno is the global marketing director of GeForce at NVIDIA. He manages a shared resource team covering social media, influencer marketing, and original content videos.
Fenno’s marketing career began at Mattel, Inc., where he managed the Hot Wheels, Batman, and DC Comics brands. He later made the leap from toys to video games, joining Electronic Arts to work on the Harry Potter, The Sims, and Maxis businesses, serving as global marketing lead for The Sims 4. Fenno’s career spans entertainment marketing, branded packaged goods, global and regional marketing, and shared service team management.
He earned a bachelor of arts from Tulane University and an M.B.A. in marketing from the University of Southern California.